Testing the Performance of CSS's Elasticsearch Vector Search
Scenarios
CSS's vector search engine provides a fully managed, high-performance distributed vector database service. To facilitate performance/pressure testing for the vector search service and provide accurate references for product selection and resource configuration, this document describes the performance testing solutions for CSS's Elasticsearch vector search service based on open-source datasets and open-source pressure testing tools.
Preparations
- Create an Elasticsearch vector database. For details, see .
Set the node quantity to 3, and node specifications to 4 vCPUs | 16GB under General computing. (The test data volume is relatively small, and the CPU specifications need to be kept close to those used in third-party performance benchmarking.) Select ultra-high I/O for node storage, and keep the security mode disabled.
- Obtain test datasets.
- sift-128-euclidean: 128 dimensions, 1 million base records, measured using the Euclidean distance.
- cohere-768-cosine: 768 dimensions, 1 million base records, measured using the cosine distance.
- gist-960-euclidean: 960 dimensions, 1 million base records, measured using the Euclidean distance.
You can download sift-128-euclidean and gist-960-euclidean at https://github.com/erikbern/ann-benchmarks. To use the cohere-768-cosine dataset, submit a service ticket.
Figure 1 Downloading sift-128-euclidean and gist-960-euclidean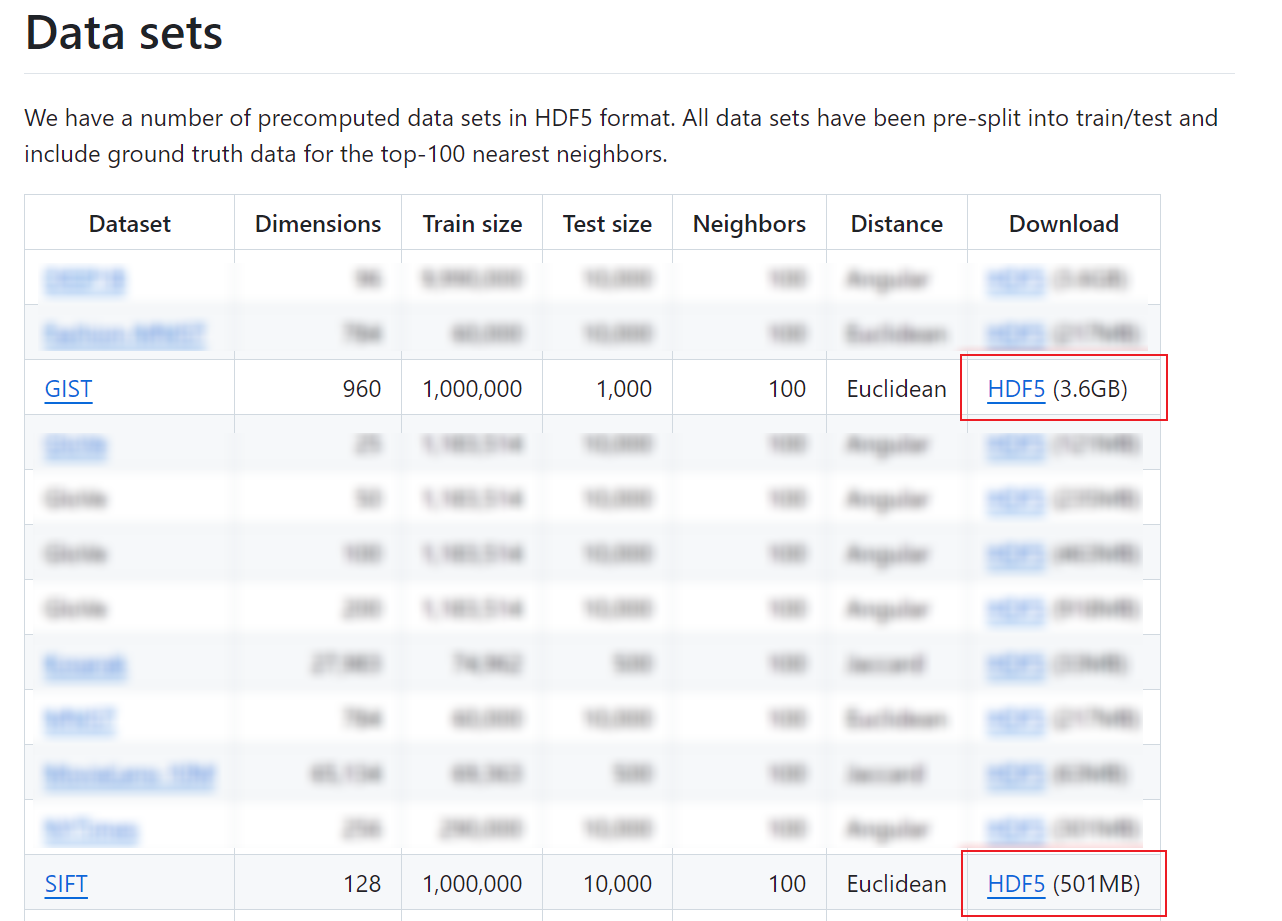
- Prepare the testing tools.
- Prepare the data writing and recall testing scripts. For details, see Script base_test_example.py.
- Download the open-source pressure testing tool wrk at https://github.com/wg/wrk/tree/master.
Performance Testing Procedure
- Create an ECS for installing the pressure testing tool and executing test scripts. For details, see .
- This ECS and the Elasticsearch cluster created for the testing purpose must be within the same VPC and security group.
- You may use another client server instead. No matter what server you use, ensure that it is in the same VPC as the Elasticsearch cluster.
- Upload the test dataset to the ECS.
- Upload the data writing and recall testing scripts to the ECS, and run the following command:
pip install h5py pip install elasticsearch==7.6 python3 base_test_example.py
This command creates a vector index for testing, writes in the test data, and returns the average recall for the queries.
- Install wrk on the ECS.
- On the ECS, prepare the query request file used for the pressure testing to simulate real-world traffic. See Script prepare_query.py for an example.
pip install h5py python3 prepare_query.py
- Prepare the wrk pressure testing configuration script on the ECS. See Script perf.lua for an example. Modify the query request file name, cluster address, and index name in the script as needed.
- Run the following command on the ECS to perform pressure testing on CSS's vector search service:
wrk -c60 -t60 -d10m -s perf.lua http://x.x.x.x:9200
- t indicates the number of pressure testing threads.
- c indicates the number of server connections.
- d indicates the pressure testing duration. 10m indicates 10 minutes.
- s indicates the pressure testing configuration script for wrk.
- x.x.x.x indicates the address of the Elasticsearch cluster.
Obtain the test result from the command output, where Requests/sec indicates the query throughput in QPS.
Figure 2 Test result example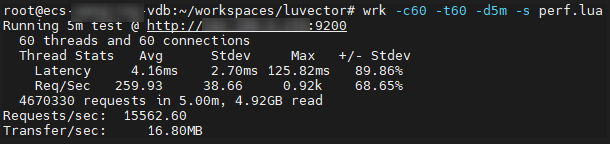
Performance Testing Solutions

The following testing solutions list only the values of the compared parameters. You are advised to retain the default values for parameters that are not listed here.
- GRAPH indexes
For a database of millions of records, GRAPH indexing is recommended.
- Testing solution 1: Use datasets of varying numbers of data dimensions to test the maximum QPS supported by the vector database when the top-10 recall rate reaches 99%. Perform the test on each dataset using both default parameters and performance-tuned ones. By tuning the build parameters, you can optimize the graph index structure to enhance query performance while maintaining the same recall rate.
Test result:
Table 1 GRAPH index test result 1 Dataset
Build Parameters
Query Parameters
Metrics
efc
shrink
ef
max_scan_num
QPS
Recall
sift-128-euclidean
200
1.0
84
10000
15562
0.99
500
0.8
50
10000
17332
0.99
cohere-768-cosine
200
1.0
154
10000
3232
0.99
500
0.95
106
10000
3821
0.99
gist-960-euclidean
200
1.0
800
19000
860
0.99
500
0.9
400
15000
1236
0.99
Conclusion: On all these datasets, the vector search service can reach a recall rate of 99% or higher using default parameters. By further tuning the build and query parameters, the index building overhead increases slightly, but this also results in improved query performance.
- Testing solution 2: Use the same dataset to test the vector search service's query performance under different recall rates by tuning index parameters. This solution uses the Cohere dataset to test the maximum QPS of the cluster under different top-10 recall rates—99%, 98%, and 95%, respectively.
Test result:
Table 2 GRAPH index test result 1 Dataset
Build Parameter
Query Parameter
Metrics
efc
ef
QPS
Recall
cohere-768-cosine
500
128
3687
0.99
500
80
5320
0.98
500
36
9028
0.95
Conclusion: With fixed index building parameters for the same cluster, tuning the ef parameter can achieve different query precisions. Slightly sacrificing the recall rate can significantly boost QPS.
- Testing solution 1: Use datasets of varying numbers of data dimensions to test the maximum QPS supported by the vector database when the top-10 recall rate reaches 99%. Perform the test on each dataset using both default parameters and performance-tuned ones. By tuning the build parameters, you can optimize the graph index structure to enhance query performance while maintaining the same recall rate.
- GRAPH_PQ indexes
Graph-based indexing typically requires residual memory to ensure query performance. When dealing with a large number of vector dimensions or high data volumes, memory resources become a crucial factor affecting costs and performance. Specifically, high-dimensional vectors and large datasets demand significantly more memory, increasing storage costs and directly impacting the efficiency and response times of the indexing algorithm. In this scenario, the GRAPH_PQ indexing algorithm is recommended.
Testing solution: Use the COHERE and GIST datasets with high data dimensions to test the cluster's maximum QPS under a top-10 recall rate of 95%. Compare the residual memory overhead with that of GRAPH indexes.
Test result:Table 3 GRAPH_PQ index test result Dataset
Build Parameters
Query Parameters
Metrics
Memory Overhead
efc
fragment_num
ef
topk
QPS
Recall
GRAPH_PQ
GRAPH
cohere-768-cosine
200
64
85
130
8723
0.95
332MB
3.3GB
gist-960-euclidean
200
120
200
360
4267
0.95
387MB
4.0GB
Conclusion: The result shows that GRAPH_PQ indexing can achieve the same or similar precision and QPS as GRAPH indexing while cutting the memory overhead more than 10 times. By integrating graph indexing and quantization, the GRAPH_PQ indexing algorithm used by CSS's vector search service significantly reduces the memory overhead and increases per-node data capacity.
Table 4 describes the indexing parameters used above. For more information about the build parameters, see . For more information about the query parameters, see .
|
Type |
Parameter |
Description |
|---|---|---|
|
Build parameter |
efc |
Queue size of the neighboring node during HNSW build. The default value is 200. A larger value indicates a higher precision and slower build speed. |
|
shrink |
Cropping coefficient during HNSW build. The default value is 1.0f. |
|
|
fragment_num |
Number of fragments. The default value is 0. The plugin automatically sets the number of fragments based on the vector length. |
|
|
Query parameter |
ef |
Queue size of the neighboring node during the query. A larger value indicates a higher query precision and slower query speed. The default value is 200. |
|
max_scan_num |
Maximum number of scanned nodes. A larger value indicates a higher query precision and slower query speed. The default value is 10000. |
|
|
topk |
The number of top-k records returned for a query. |
Script base_test_example.py
# -*- coding: UTF-8 -*-
import json
import time
import h5py
from elasticsearch import Elasticsearch
from elasticsearch import helpers
def get_client(hosts: list, user: str = None, password: str = None):
if user and password:
return Elasticsearch(hosts, http_auth=(user, password), verify_certs=False, ssl_show_warn=False)
else:
return Elasticsearch(hosts)
# For more information about the index parameters, see .
def create(es_client, index_name, shards, replicas, dim, algorithm="GRAPH",
metric="euclidean", neighbors=64, efc=200, shrink=1.0):
index_mapping = {
"settings": {
"index": {
"vector": True
},
"number_of_shards": shards,
"number_of_replicas": replicas,
},
"mappings": {
"properties": {
"id": {
"type": "integer"
},
"vec": {
"type": "vector",
"indexing": True,
"dimension": dim,
"algorithm": algorithm,
"metric": metric,
"neighbors": neighbors,
"efc": efc,
"shrink": shrink,
}
}
}
}
es_client.indices.create(index=index_name, body=index_mapping)
print(f"Create index success! Index name: {index_name}")
def write(es_client, index_name, vectors, bulk_size=1000):
print("Start write! Index name: " + index_name)
start = time.time()
for i in range(0, len(vectors), bulk_size):
actions = [{
"_index": index_name,
"id": i + j,
"vec": v.tolist()
} for j, v in enumerate(vectors[i: i + bulk_size])]
helpers.bulk(es_client, actions, request_timeout=180)
print(f"Write success! Docs count: {len(vectors)}, total cost: {time.time() - start:.2f} seconds")
merge(es_client, index_name)
def merge(es_client, index_name, seg_cnt=1):
print(f"Start merge! Index name: {index_name}")
start = time.time()
es_client.indices.forcemerge(index=index_name, max_num_segments=seg_cnt, request_timeout=7200)
print(f"Merge success! Total cost: {time.time() - start:.2f} seconds")
# For more information about the query parameters, see .
def query(es_client, index_name, queries, gts, size=10, k=10, ef=200, msn=10000):
print("Start query! Index name: " + index_name)
i = 0
precision = []
for vec in queries:
hits = set()
dsl = {
"size": size,
"stored_fields": ["_none_"],
"docvalue_fields": ["id"],
"query": {
"vector": {
"vec": {
"vector": vec.tolist(),
"topk": k,
"ef": ef,
"max_scan_num": msn
}
}
}
}
res = es_client.search(index=index_name, body=json.dumps(dsl))
for hit in res['hits']['hits']:
hits.add(int(hit['fields']['id'][0]))
precision.append(len(hits.intersection(set(gts[i, :size]))) / size)
i += 1
print(f"Query complete! Average precision: {sum(precision) / len(precision)}")
def load_test_data(src):
hdf5_file = h5py.File(src, "r")
base_vectors = hdf5_file["train"]
query_vectors = hdf5_file["test"]
ground_truths = hdf5_file["neighbors"]
return base_vectors, query_vectors, ground_truths
def test_sift(es_client):
index_name = "index_sift_graph"
vectors, queries, gts = load_test_data(r"sift-128-euclidean.hdf5")
# Adjust the number of shards and replicas, indexing algorithm, and index parameters based on the testing requirements. In our example here, one shard and two replicas are configured for performance testing.
create(es_client, index_name, shards=1, replicas=2, dim=128)
write(es_client, index_name, vectors)
query(es_client, index_name, queries, gts)
if __name__ == "__main__":
# Change the value to the address of the CSS cluster.
client = get_client(['http://x.x.x.x:9200'])
test_sift(client)
Script prepare_query.py
import base64
import json
import struct
import h5py
def prepare_query(src, dst, size=10, k=10, ef=200, msn=10000, metric="euclidean", rescore=False, use_base64=True):
"""
This function is used to read query vectors from the source data files in HDF5 format and generate a complete query request body for performance testing.
:param src: path of the source data files in HDF5 format.
:param dst: destination file path.
:param size: number of query results returned.
:param k: number of top-k similar results returned by querying a segment-level index.
:param ef: specifies the queue size used during a query.
:param msn: specifies max_scan_num.
:param metric: metric used for result rescoring, such as euclidean, cosine, and inner_product.
:param rescore: whether to use rescoring. You can enable rescoring for GRAPH_PQ indexes.
:param use_base64: whether to use Base64-encoded vector data.
"""
hdf5_file = h5py.File(src, "r")
query_vectors = hdf5_file["test"]
with open(dst, "w", encoding="utf8") as fw:
for vec in query_vectors:
query_template = {
"size": size,
"stored_fields": ["_none_"],
"docvalue_fields": ["id"],
"query": {
"vector": {
"vec": {
"vector": vec.tolist() if not use_base64 else floats2base64(vec),
"topk": k,
"ef": ef,
"max_scan_num": msn,
}
}
}
}
if rescore:
query_template["query"]["rescore"] = {
"window_size": k,
"vector_rescore": {
"field": "vec",
"vector": vec.tolist() if not use_base64 else floats2base64(vec),
"metric": metric
}
}
fw.write(json.dumps(query_template))
fw.write("\n")
def floats2base64(vector):
data = struct.pack('<{}f'.format(len(vector)), *vector)
return base64.b64encode(data).decode()
if __name__ == "__main__":
# Change the value to the data file address.
prepare_query(r"/path/to/sift-128-euclidean.hdf5", r"requests.txt")
Script perf.lua
local random = math.random
local reqs = {}
local cnt = 0
-- Rename the query request file used for pressure testing as needed.
for line in io.lines("requests.txt") do
table.insert(reqs, line)
cnt = cnt + 1
end
local addrs = {}
local counter = 0
function setup(thread)
local append = function(host, port)
for i, addr in ipairs(wrk.lookup(host, port)) do
if wrk.connect(addr) then
addrs[#addrs+1] = addr
end
end
end
if #addrs == 0 then
-- Change the value to the cluster address.
append("x.x.x.x", 9200)
append("x.x.x.x", 9200)
append("x.x.x.x", 9200)
end
local index = counter % #addrs + 1
counter = counter + 1
thread.addr = addrs[index]
end
-- Change the index name as needed.
wrk.path = "/index_sift_graph/_search?request_cache=false&preference=_local"
wrk.method = "GET"
wrk.headers["Content-Type"] = "application/json"
function request()
return wrk.format(wrk.method, wrk.path, wrk.headers, reqs[random(cnt)])
end
Feedback
Was this page helpful?
Provide feedbackThank you very much for your feedback. We will continue working to improve the documentation.






